Abstract
Multiple Myeloma (MM) is an incurable malignancy of bone marrow plasma cells characterized by wide molecular diversity and complex clonal and subclonal architecture. Intra-tumor heterogeneity reflects the evolution of the disease. Clonal structure may affect response to treatment, which, in turn, may shape tumor evolution and, consequently, contribute to drug resistance. Thus, analysis of tumor clonality has profound implications for personalized medicine.
Using our recently developed network model of newly diagnosed MM from the MMRF CoMMpass study (Lagana et al, Leukemia 2017) , MMNet, we have identified patterns of gene co-expression significantly associated with tumor clonality and showed that clonality is correlated with mutational burden and relapse.
In order to characterize the evolution of clonal structure from diagnosis to relapse, we analyzed Whole-Exome data (WES) from 19 patients enrolled in CoMMpass for whom sequencing data for multiple time points was available. The cohort included both male (12; 63%) and female (7; 37%) patients of white/caucasian (14; 73%) and black/african-american (5; 27%) origin. Patients were distributed among five of the ten classes defined by MMNet: CCND1 (4; 21%), MAF (1; 5%), MMSET (2; 10%), HY/NRAS (4; 21%), and IMM (4; 21%) (4 were unassigned). Also, six patients had the t(11;14) and two had the t(4;14) chromosomal translocations. All patients had received induction therapy, consisting of combinations of proteasome inhibitors (PI) (Bortezomib: 19, 100%; Carfilzomib: 3, 16%), immunomodulatory drugs (IMiDs) (Lenalidomide: 13, 68%; Pomalidomide: 2, 10%; Thalidomide: 1, 5%), steroids (18, 95%) and monoclonal antibodies (Daratumumab: 1, 5%). Best responses to therapy were CR/sCR (3; 16%), VGPR (7; 37%) and PR (9; 47%). Fourteen (68%) patients had SD or PD at the second sequencing time point, with a median PFS of 456 days.
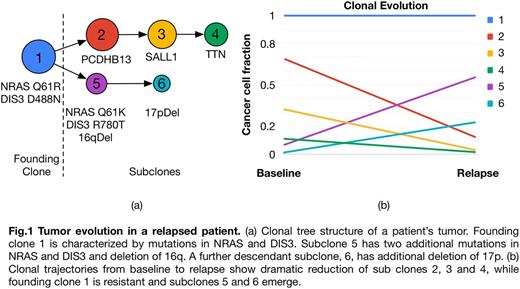
Our analysis inferred subclonal tumor composition and evolution based on somatic mutations and copy number alterations (CNA) using PhyloWGS (Deshwar et al, Genome Biol 2015). Patients had one (14; 74%), two (4; 21%) or three (1; 5%) founding clones at baseline, and numerous subclones. Trajectories of cancer cell populations revealed dramatic changes in most patients in terms of clonal and subclonal cell fractions between baseline and relapse. In most patients we observed emergence of new competing clones at relapse, where one or more subclones decreasing in size were replaced by other subclones, indicating selective pressure introduced by therapy (Fig. 1).
Subclones were characterized in terms of mutations and CNA and were labeled as stable/resistant or sensitive based on their trajectories from baseline to relapse. Our analysis revealed significant inter-patient heterogeneity in terms of mutations and clonal composition, with few overlaps. We identified subclonal drivers by screening the observed mutations and CNA against a database of known cancer drivers and analysis of co-occurrence and mutual exclusivity. Our analysis revealed that stable/resistant clones were characterized by concurrent deletion of 17p and 13q, and/or mutations in NRAS. In particular, 7 out of 19 patients had at least one mutation in NRAS, observed at baseline in 5 cases, and with two patients carrying two different mutations in two different clones. Our findings support earlier adoption of targeted therapy against RAS (e.g. Trametinib). Other drivers found exclusively in resistant clones included DIS3, FAM46C, ROBO1 and CCND1.
Overall, our analysis provides genomics characterization of relapsed patients in CoMMpass following induction therapy, reveals heterogeneous clonal and subclonal composition and trajectories from baseline to relapse, and defines specific somatic mutations (e.g. NRAS) and CNA as drivers of resistance to induction therapy.
Madduri: Foundation Medicine, Inc.: Consultancy. Chari: Janssen: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Array BioPharma: Consultancy, Research Funding; Millennium Pharmaceuticals, Inc.: Consultancy, Research Funding; Amgen: Honoraria, Research Funding; Celgene Corporation: Consultancy, Research Funding. Cho: Bristol Myers-Squibb: Other: advisory board, Research Funding; Agenus, Inc.: Research Funding; Genentech: Other: advisory board, Research Funding; Ludwig Institute for Cancer Research: Research Funding; Multiple Myeloma Research Foundation: Research Funding. Barlogie: Millenium Pharmaceuticals: Consultancy, Research Funding; Celgene Corporation: Consultancy, Research Funding. Jagannath: Celgene: Consultancy; Bristol-Meyers Squibb: Consultancy; Merck: Consultancy; Medicom: Speakers Bureau; MMRF: Speakers Bureau; Novartis: Consultancy. Dudley: GlaxoSmithKline: Consultancy; Janssen Pharmaceuticals, Inc.: Consultancy; Ayasdi, Inc.: Equity Ownership; Ecoeos, Inc.: Equity Ownership; NuMedii, Inc.: Equity Ownership, Patents & Royalties; Ontomics, Inc.: Equity Ownership; Personalis: Patents & Royalties; AstraZeneca: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal